Scientists have developed a new platform called nELISA that can measure hundreds of proteins simultaneously from thousands of biological samples with unprecedented speed and accuracy. This technology addresses a major bottleneck in medical research, potentially accelerating the discovery of new drugs and diagnostic tools for a wide range of diseases.
The platform combines a novel immunoassay design with advanced bead-barcoding technology, allowing researchers to generate roughly 1.4 million protein measurements in under a week. This high-throughput capability opens new avenues for understanding complex biological systems and immune responses.
Key Takeaways
- A new technology called nELISA has been developed for high-throughput, multiplexed protein analysis.
- It overcomes a key limitation of traditional methods called reagent-driven cross-reactivity (rCR), allowing for a 191-protein panel to be run simultaneously.
- The system can process thousands of samples per week, achieving a sensitivity down to sub-picogram-per-milliliter levels.
- In a demonstration, nELISA successfully profiled 7,392 samples, revealing over 440 distinct cytokine responses and uncovering new biological insights.
- The technology shows strong potential to accelerate drug discovery, biomarker identification, and personalized medicine.
The Challenge of Protein Profiling
Understanding the role of proteins is fundamental to biology and medicine. Unlike genomics, which studies our genetic blueprint, proteomics examines the proteins that perform most of the work inside our cells. However, accurately measuring a wide array of proteins at once has been a significant technical hurdle.
Traditional methods like the enzyme-linked immunosorbent assay (ELISA) are highly accurate but are typically limited to measuring one protein at a time. Attempts to scale up these assays often run into a problem called reagent-driven cross-reactivity (rCR). When antibodies for different proteins are mixed, they can mistakenly interact, creating false signals and compromising data quality. This has historically limited multiplexed immunoassays to panels of around 25 proteins or fewer.
Why Direct Protein Measurement Matters
While genetic and RNA analysis provide valuable information, they don't tell the whole story. Protein levels are influenced by complex regulatory processes that occur after a gene is transcribed. Direct protein measurement is essential for a true understanding of cellular function, disease states, and a patient's response to treatment.
Introducing the nELISA Platform
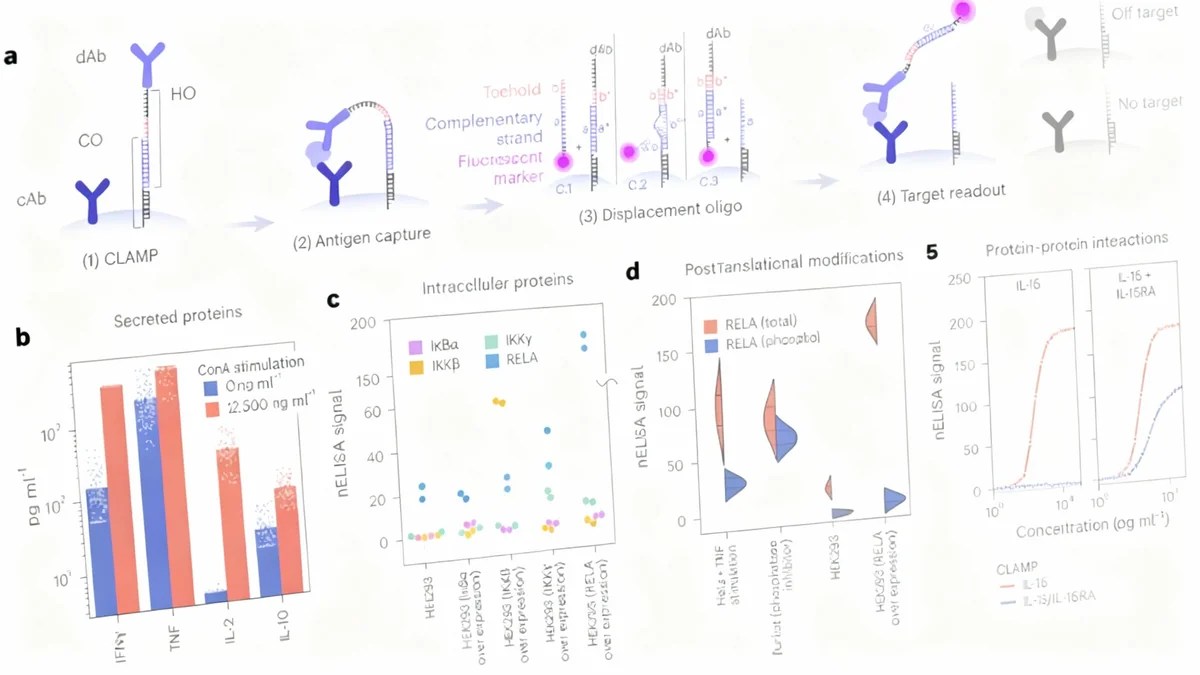
The newly developed nELISA platform introduces a groundbreaking approach to solve the cross-reactivity problem. It is built on two core innovations: a novel assay design called CLAMP and a sophisticated bead-encoding system.
CLAMP: Preventing Unwanted Interactions
The CLAMP (colocalized-by-linkage assays on microparticles) system re-engineers the traditional sandwich immunoassay. Instead of mixing antibodies freely in a solution, each antibody pair is pre-assembled and confined to the surface of a microscopic bead. This spatial separation physically prevents non-cognate antibodies from interacting, effectively eliminating the primary source of cross-reactivity.
The detection process is also unique. It uses a DNA-based mechanism where a fluorescently labeled molecule displaces and labels the detection antibody only when the target protein is present. If the target protein is absent, the fluorescent probe is washed away, resulting in a very low background signal and high sensitivity.
Exceptional Specificity: In a test involving over 36,000 potential non-cognate interactions within a 191-protein panel, only five cross-reactive events were detected, three of which were due to known biological similarities between proteins.
High-Throughput Barcoding
To analyze hundreds of proteins at once, each bead targeting a specific protein is given a unique spectral "barcode." This is achieved using a technique called emFRET, which combines four standard fluorophores in varying ratios to create thousands of distinct signatures. A flow cytometer can then rapidly read the barcode of each bead and measure the fluorescent signal associated with the target protein.
This combination allows a single, pooled sample of barcoded beads to be added to a patient sample, with the results for all 191 proteins read out in a single, rapid analysis. The workflow is optimized for a 384-well plate format, enabling the profiling of 1,536 wells per day on a single cytometer.
Performance and Validation
The nELISA platform has demonstrated impressive performance, matching or exceeding the capabilities of existing technologies. Its sensitivity allows for the detection of proteins at concentrations as low as 0.1 picograms per milliliter (pg/ml).
Furthermore, the system provides a wide dynamic range, capable of quantifying proteins across seven orders of magnitude. This is crucial for biological samples where protein concentrations can vary dramatically. Reproducibility is also high, with coefficients of variation around 3% for measurements within the same plate and 5% between plates run on different days.
When compared to established multiplex platforms like xMAP and Olink's PEA, nELISA showed strong agreement. For 36 shared targets with the xMAP platform, the median Spearman correlation was 0.92. Against the Olink platform, the median correlation for shared targets was above 0.95.
Application in Immune Response Screening
To showcase its real-world utility, researchers used the 191-plex inflammation panel to conduct a large-scale functional screen of peripheral blood mononuclear cells (PBMCs), a key component of the human immune system. PBMCs from six donors were stimulated with various inflammatory agents and 80 different immunomodulatory proteins.
The screen generated 7,392 high-content protein profiles, a dataset of nearly 1.4 million individual measurements. The results clearly clustered based on the type of stimulus, donor, and concentration, demonstrating the platform's ability to capture nuanced biological responses.
The data not only recapitulated known immune pathways but also revealed novel insights:
- It captured the suppressive effects of anti-inflammatory cytokines like IL-4 and IL-10 on protein expression, effects often missed by transcriptomic (mRNA-based) methods.
- It highlighted the role of chemokines in modulating cytokine secretion, a function beyond their traditional role in cell migration.
- The platform identified functional similarities between therapeutic drugs, such as IFNβ and IL-1RA (anakinra), providing data that could inform drug repurposing for conditions like multiple sclerosis.
Future Implications for Medicine
The development of nELISA represents a significant step forward for proteomics. By providing a cost-effective, scalable, and highly accurate method for protein profiling, it bridges a critical gap between genomics and functional biology.
Its compatibility with other cell-based assays, such as high-content imaging (Cell Painting), further enhances its value. By combining morphological data with secretome data from the same sample, researchers can gain a more complete mechanistic understanding of a drug's effect.
This technology is poised to have a broad impact on various fields, including drug discovery, biomarker identification, immune monitoring, and clinical diagnostics. The ability to rapidly and affordably analyze the protein landscape of thousands of samples could fundamentally change how we study disease and develop new therapies.